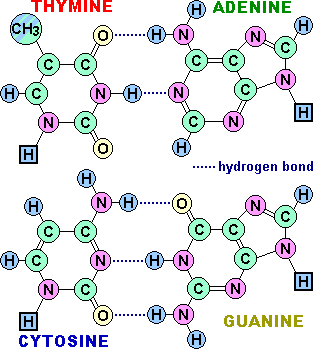
The simple and inflexible rules of base pairing tell us that if we can "read" the sequence of nucleotides on one strand of DNA, we can immediately infer the complementary sequence on the other strand.
The terms DNA, Chromosomes and Genes are often used interchangeably, but they are not just different terms for the same thing. The tangle of protein fibres in the nucleus of a cell when it is not actively dividing is called chromatin and has the general term DNA.
At cell division the chromatin separates into chromosomes. Each chromosome contains an individual DNA molecule, and each DNA molecule is made up of many individual segments known as genes. Each gene in turn is a sequence of locations known as loci that can be occupied by alleles. Alleles themselves are any one of a number of possible short sequences of DNA coding that can occupy a given locus or position on the strand of DNA. A diploid organism like the honey bee has two copies of each chromosome, the two alleles at each corresponding locus make up the individual's genotype.
Genes can be sequenced at any level the total set being known as the genome, but as the amount of data is huge, it is usually expressed as shorter sequences that are mapped to each other by position on a chromosome or within a gene segment.
A human being has 23 pairs, making 46 chromosomes in all, our honey bees have 32 chromosomes organised as 16 pairs.
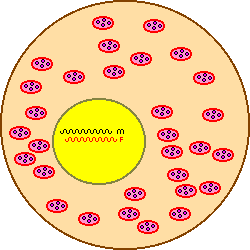
Nuclear DNA in diploid organisms (female bees) has two copies... One is inherited from each parent.
Mitochondrial DNA (Mt DNA) is inherited through the maternal line only, without recombination, and so is not unique to an individual, but represents the whole line or family. It can be used to examine the degree of relatedness and common ancestry between individuals in a population, in the case of our honey bees this is a feature of the queen of a colony and can be used to trace pedigrees of strains and has many inferences in the origin and evolution of strains and races. Many copies of the Mitochondrial DNA are present in each cell (maybe as many as a thousand). The illustration at right shows male and female copies of parental DNA within the nucleus with the mitochondria floating in the cytoplasm. The complete sequence of honeybee (Apis mellifera) mitochondrial DNA is reported as being 16,343 bp in length.
Minisatellite DNA (sometimes referred to as a variable number of tandem repeats or VNTRs) contains medium length segments from 1,000 to 20,000 base pairs. The repeated unit ranges from 9 to 80 base pairs and they
occur in non-coding regions.
Microsatellite DNA (sometimes referred to as a short tandem repeats or STRs) are short segments of DNA that have a repetitive character in their sequence such as CACACACA... This can be represented as (CA)4, these repeated
regions tend to occur in non-coding parts of the DNA. The repeat unit consists of 1 to 6 base pairs and the whole repeated section is less than 150 base pairs.
The DNA sequences that encode the most important information are known as "Coding DNA". The parts of a DNA sequence that do not encode anything (or at least as far as our current knowledge) are referred to as
"Non-coding DNA" (sometimes just known as "Junk DNA").
Each of the ends of a strand of DNA will have a polarity that will be opposite from the polarity of the other end. One end of the DNA strand is referred to as the 5-prime end, and the other end is referred to as the
3-prime end. During the amplification process, DNA is synthesized from the free 3-prime ends of a pair of single-stranded DNA. These small segments of DNA are known as the "primers" The primers are complimentary to one of the sides of the double-stranded DNA that is to be amplified and will thus "prime" the amplification reaction. So a single pair of PCR primers will produce different sized products for each of the different length microsatellites.
The polymerase chain reaction (PCR) can be used to make many copies (amplification) of specific regions or fragments of DNA. By splitting the two strands apart to form template strands in a 'soup' of spare bases and repeatedly raising and lowering the temperature, the spare bases latch on to their opposing types and each single strand becomes a double one again. Each cycle that this reaction goes through, doubles the amount of DNA present. After thirty or forty cycles, many millions of copies of the specified region of DNA will be present.
This gives us a way to detect microsatellites by finding regions that are unique to one locus in the genome that have unique structure on either side of the repeated portion.
Gel Electrophoresis... Agarose or polyacrylamide gels have channels within their matrix structure that are of similar size to molecules. Since nucleic acids are negatively charged, we can apply an electric field to a gel
that has samples of amplified DNA and they will migrate towards the positive pole of the applied voltage. As shorter chains move faster than longer ones the different sized molecules are spread out on the gel in an inverse relationship to their size.
PCR products can be prepared for DNA sequencing by another method... The dideoxy terminator method, also known as Sanger's Method, is used for Mt DNA cycle sequencing. The cycle sequencing process is similar to the PCR, but different chemicals are used. A set of terminator bases are used in addition to the normal bases that elongate the growing strand of DNA. These terminator bases lack a chemical group that would normally allow the enzyme to place another base after them. The altered bases also carry a fluorescent dye can be detected by an a fluorescence detector that records the emitted wavelength of the fluorescent dyes on each base as the fragments travel past
the detection area of the instrument. The instrument produces a chromatogram, the colours of which mimic the labeled fragments. The normal bases compete with the altered bases for incorporation into the growing DNA strand, resulting in a collection of DNA products that differ in size by one base and have a fluorescently labeled
base at their end.
The term "polymorphism" describes the existence of different forms within a population, a difference in the number of tandem repeats between close relations gives rise to multiple forms of an allele within in a population.
The Apis mellifera genome has high A+T and CpG contents and is approximately 200M bases in length.
Dave Cushman.
Page created pre-2011
Page updated 18/12/2022